|
Congrats to the freshman biology students in my Introductory Biology lab section (Biol126) on their recent publication! Students in the class isolated bacteria from soil samples from Geneseo, NY, classified them, and tested their ability to produce antibiotics under different conditions. They found three strains that inhibited the growth of Neisseria commensals, close relatives of important human pathogens. We ultimately hope that these compounds will also inhibit the growth of gonococcal strains, providing potential new antimicrobial therapies for treatment of gonorrhea.
https://journals.asm.org/doi/10.1128/mra.00627-22
0 Comments
I am so excited to see this work finally published! This study was designed and conducted by the students in my Genomics class at RIT during the fall semester of 2020. In this work, students experimentally evolved resistance to an antibiotic in a Neisseria commensal species. They then sequenced the genomes of resistant lineages and found mutations that caused resistance converged on short sequence duplication events in ribosomal genes. Since Neisseria commensals share resistance with their pathogenic relatives (one of which is the bacterial pathogen that causes gonorrhea), it is important to understand the paths to resistance in these species too!
Congrats to all involved! An especially big thank you to André O. Hudson for your help and guidance! https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0262370 A big congratulations to RIT students Michael Foire and Jordan Raisman on their new preprint (https://www.biorxiv.org/content/10.1101/2020.07.30.228593v1)! This work characterizes resistance across the commensal Neisseria, which are known sources of resistance for their pathogenic cousin, Neisseria gonorrhoeae. Using both MIC testing and whole genome sequencing, several resistance-encoding mutations are nominated in non-pathogenic species.
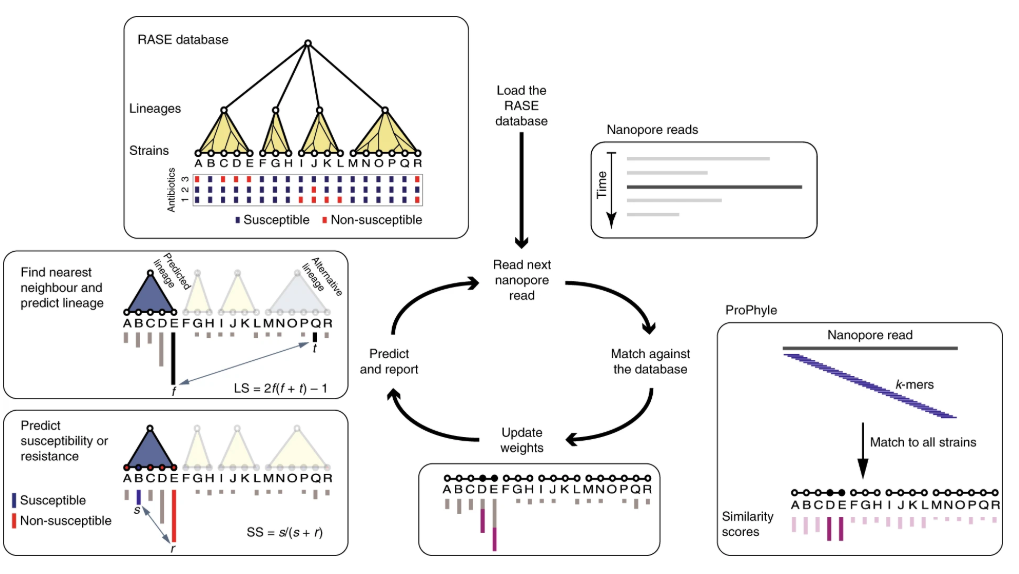
Congratulations to collaborators on the publication of "Rapid inference of antibiotic resistance and susceptibility by genomic neighbor typing" in Nature Microbiology! Here we present a new method for identifying resistant bacteria within minutes (<10 min), using real-time Nanopore sequencing and phenotypic inference from near phylogenetic neighbors. One step closer to point of care resistance diagnostics which will enable tailored therapies for individual patients! Check it out here: https://doi.org/10.1038/s41564-019-0656-6.
The lab is finally coming together, and I am so excited to get started with my incoming students this semester. I can not wait to see the paths we will take towards discovery, the unexpected challenges and creative solutions used to overcome these obstacles, and the team that we will build. Here's to the future, watch out gonococcus!
New Preprint: Using Linkage Patterns to Understand the Selective Pressures That Bacteria Face5/15/2019 The ecological pressures that bacteria face can be hard to determine, however selection leaves clear signatures within the genome. In our recent preprint, spearheaded by Dr. Brian Arnold, we look at patterns of linkage to nominate the selective pressures faced by the recombining bacterial pathogen Neisseria gonorrhoeae. Clear signatures emerge, suggestive of a large role of adaptive interspecies admixture and/or balancing selection (perhaps due to the existence of distinct ecological niches or colonization sites) in shaping population level diversity in this species. Congratulations to all involved!
See our preprint here: https://www.biorxiv.org/content/10.1101/634147v1 Our study, lead by Dr. Genevieve M. Kozak, was recently published as a preprint on bioRxiv. Congratulations to all involved! This work highlights that the genetic basis of complex traits like seasonal timing and resultant ecological speciation, can be controlled by a relatively simple genetic basis. Here, we find that two genes involved in the circadian clock pathway underlie this trait in the European Corn Borer moth, a major agricultural pest of corn in North America and Europe. Ultimately, describing the genetic basis of seasonality may help us better predict the extinction or survival outcomes due to climate change for particular species, and also gain a better understanding of range expansions in insect systems.
See our preprint here: https://www.biorxiv.org/content/10.1101/633362v1
|
AuthorCrista Wadsworth is an Assistant Professor at RIT. Current projects focus on the resistome within commensal Neisseria. Archives
September 2022
Categories |









 RSS Feed
RSS Feed